A02
Chloroplast Ribonucleoprotein CP29A – stabilizing chloroplast RNA pools during cold acclimation responses
The chloroplast RNA binding protein CP29A associates with multiple RNAs and is required for their stability during cold acclimation. We determined its RNA binding sites at base resolution and found its main target on the rbcL mRNA, next to the PPR protein MRL1, which is known to support RNA stability and translation of rbcL. CP29A has a prion-like domain and we demonstrate that it is capable of liquid-liquid phase separation (LLPS) in vitro and is found in cold-dependent granules in vivo. Our goal is to determine the role of LLPS and CP29A-MRL1 interactions for the expression of rbcL in the cold using a combination of genetic, biochemical, and super-resolution micros-copy approaches.

Cold-dependent phenotypes and expression of CP31A and CP29A: Phenotype of plants grown for 2 weeks at 23°C and subsequently 5 weeks at 8°C. The center of the rosette bleaches.
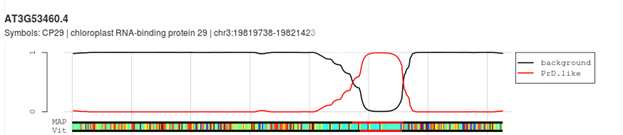
CP29A has a prion-like domain: A bioinformatics analysis of CP29A using the PLAAC tool to screen for foldedness uncovers the presence of a highly disordered, prion-like domain (PLD; LCR = low complexity region) sandwiched between the two RMM domains.

